|
COORDINATOR: PAOLA BONFANTE
The aim of the research project is to provide the description of the complete genome sequence of the bacterial endosymbiont Candidatus Glomeribacter gigasporarum which lives inside the arbuscular mycorrhizal fungus Gigaspora margarita which lives associated to the root plants.
Symbiotic associations between endocellular bacteria and eukaryotic cells are widespread among animals and plants, but only a few examples have been described in the fungal kingdom. Arbuscular mycorrhizal (AM) species, belonging to the family Gigasporaceae, represent a specialized niche for rod shaped bacteria, named Candidatus Glomeribacter gigasporarum (Bianciotto et al, 2003). Morphological and molecular studies on G. margarita BEG34 have shown that Ca. G. gigasporarum is a homogeneous population closely related to the b-proteobacterial genus Burkholderia. Ca. G. gigasporarum is vertically transmitted through fungal generations and all attempts to grow it in pure culture failed (Bianciotto et al, 2004). Candidatus has a relatively small genome, approximately 1.4Mb (Jargeat et al, 2004). These findings suggest that these bacteria are obligate endocellular components of their AM fungal host.
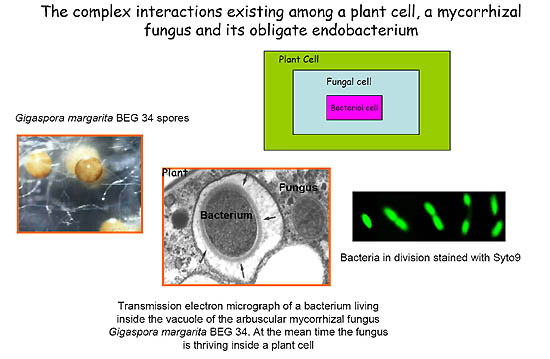
Bacteria living inside AM fungi represent thus far a hidden microbial world for which almost no genomic information is currently available despite their potentially important impacts on their eukaryotic hosts. Because Ca. G. gigasporarum can not be cultured apart outside of the host for traditional genetic or physiological techniques, analysis of the genome sequences offers a valuable tool to infer its metabolic functions. The complete genome sequence of Ca. G. gigasporarum will provide a critical step forward in our ability to answer fundamental questions concerning the biology, ecology and evolutionary history of this endosymbiont. In addition, this information may have a relevant role also for applicative purposes. Since the pioneer work of Craig Venter (2004) on the Sargasso sea, an emerging field of research is the exploration of the genetic diversity of unculturable microorganisms from environmental samples, through the so called Metagenomic approach, with the aim to discover new bioactive molecules of biotechnological interest. For this reason metagenomes have attracted the attention of academia and private companies (Lorenz & Eck, 2005).
The main aim of the WP is to provide an overall genomic information of Candidatus Glomeribacter gigasporarum by using a metagenomic approach. In collaboration with Peter Lammers (New Mexico State University) we have constructed a genomic library into a fosmid vector starting from the fungal spores (Gigaspora margarita). The library has been demonstrated to be representative of the Ca. G. gigasporarum genome.
For more than twenty years the corner stone of whole genome sequencing was the Sanger method that suffered modifications and variations with substantial improvement in the sequencing techniques and data analysis. The Sanger method is still in the top of the sequencing methods even today due to its quality to produce long reads of DNA sequences, but recently new methods such as ‘pyrosequencing’, which is at the basis of the ‘454 technology’, are becoming popular as complementary methods that allow sequencing of difficult genomic regions and improve the overall reading coverage (Goldberg et al 2006).
According to this current situation, we will face the two experimental strategies:
1)The full genome sequencing approach, based on the Sanger sequencing technology, will be performed on validated clones (current number of validated clones: 1095). The sequencing will be done in collaboration with the Parco Tecnologico padano, PTP http://www.tecnoparco.org
2)454 technology which will be applied on a purified bacterial DNA fraction, which will require the development of new protocols (bacteria isolation and whole genome amplification).
|

